Setting up a continuous integration pipeline for an R Bioconductor package with Travis
I’ve been thinking about setting up a continuous integration pipeline for the chimeraviz package for a while. Two things held me back:
- I thought it would be a hassle to make work.
- I was the only contributor, so I could just build it on my own machine.
Since I’ve recently received several pull requests to chimeraviz, the first point is no longer true, so I thought I’d finally challenge the second point.
What is Travis?
Travis CI is a continuous integration service used to build and test open source projects hosted on GitHub. Setting up R projects with Travis used to be a hassle, but now Travis has native support for the R language: https://docs.travis-ci.com/user/languages/r/. The best thing about it? It’s free when your repository is public. Since I have a GitHub repository for chimeraviz in addition to the Bioconductor git repository, Travis is a great fit.
Using Travis to build chimeraviz
To enable Travis builds for chimeraviz, I first had to sign in on Travis using my GitHub account. Once signed in, I simply had to flick a switch to enable Travis builds for my chimeraviz repository.
For Travis to know how to build chimeraviz, I had to create a .travis.yml configuration file. The getting started site for R had a few pointers, and after a few tries I got it working with this:
# R for travis: see documentation at
# https://docs.travis-ci.com/user/languages/r
language: r
sudo: false
cache: packages
r:
- bioc-devel
warnings_are_errors: true
script:
- Rscript -e "library(devtools); devtools::check()"
notifications:
email:
on_success: always
on_failure: always
Note this part:
r:
- bioc-devel
Here I specify which version of R should be used for my build. Luckily, Travis has R versions including Bioconductor for both its release (bioc-release) and development (bioc-devel) versions, so I don’t have to do anything special to get Bioconductor packages installed.
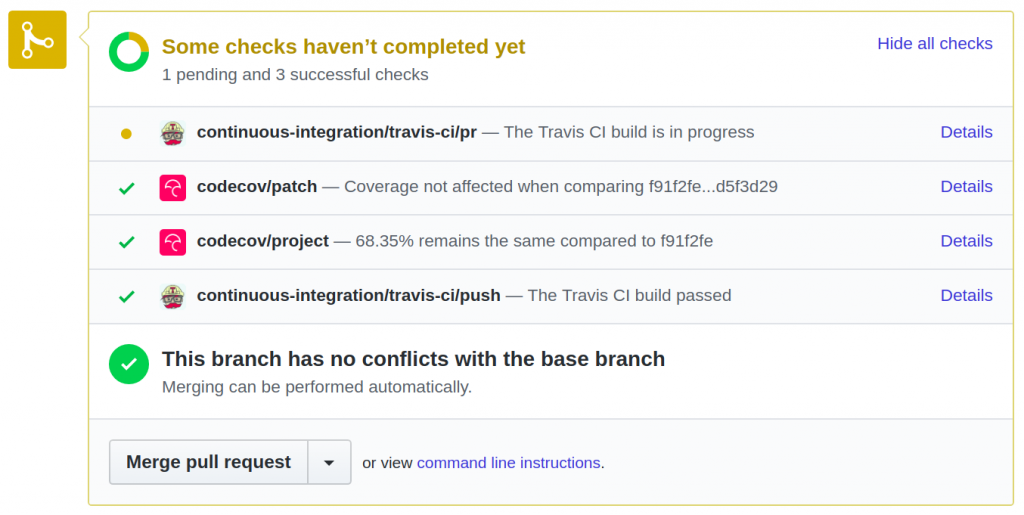
With this in place, contributors submitting pull requests to chimeraviz will get feedback on whether their changes introduce any problems with the build:
The next step is to introduce the use of a code coverage tool. I’ll update this post with the additional information when I get that working as well.
UPDATE: Adding code coverage report with codecov.io was very easy - I just had to add this to the .travis.yml file:
script:
- Rscript -e "library(devtools); devtools::check()"
r_github_packages:
- r-lib/covr
The complete .travis.yml then looks like this:
# R for travis: see documentation at https://docs.travis-ci.com/user/languages/r
language: r
sudo: false
cache: packages
r:
- bioc-devel
warnings_are_errors: true
script:
- Rscript -e "library(devtools); devtools::check()"
r_github_packages:
- r-lib/covr
after_success:
- Rscript -e 'covr::codecov()'
notifications:
email:
on_success: always
on_failure: always
And I’m able to show the code coverage on the README of the package:
As well as have these checks run automagically whenever someone submits a pull request: